MEA definition¶
This notebook shows how MEA can be using a .yaml file and how MEA models can be added and removed to and from the file system.
List available MEAs:¶
MEA.return_mea()
Available MEA:
['SqMEA-6-25um', 'SqMEA-10-15um', 'tetrode', 'Neuroseeker-128', 'SqMEA-5-30um', 'SqMEA-15-10um', 'Neuronexus-32-Kampff', 'Neuronexus-32-cut-30', 'Neuropixels-128', 'Neuroseeker-128-Kampff', 'Neuropixels-24', 'SqMEA-7-20um', 'Neuronexus-32', 'Neuropixels-384']
These MEA are saved during installation. Each MEA corresponds to a .yaml file containing key information for the MEA. Let’s take a look at some examples.
Square MEA¶
sqmea_info = MEA.return_mea_info('SqMEA-10-15um')
pprint(sqmea_info)
{'dim': 10,
'electrode_name': 'SqMEA-10-15um',
'pitch': 15,
'shape': 'square',
'size': 5,
'sortlist': None,
'type': 'mea'}
The returned dictionary corresponds the the .yaml file. For this MEA
model dim is a single int and pitch is a single int (or
float). Therefore, a 10x10 Square MEA is instantiated with 15um
pitch in the yz direction (if plane is not in the yaml file, yz
is default). The electrodes shape is square, and half the side
length is 5um. Since sortlist is None, the electrode count
starts from the bottom left and it follows the rows up and then goes to
the next column (the last index is the electrode on the top right). The
type mea will be used for plotting.
Let’s now instantiate a MEA object:
sqmea = MEA.return_mea('SqMEA-10-15um')
print(type(sqmea))
print(sqmea.number_electrodes)
print(sqmea.dim)
'plane' field with 2D dimensions assumed to be 'yz
Model is set to semi
<class 'MEAutility.core.RectMEA'>
100
[10, 10]
The MEA is a rectangular MEA with 100 electrodes.
plt.plot(sqmea.positions[:, 1], sqmea.positions[:, 2], 'b*')
plt.plot(sqmea.positions[0, 1], sqmea.positions[0, 2], 'r*')
plt.plot(sqmea.positions[9, 1], sqmea.positions[9, 2], 'g*')
plt.plot(sqmea.positions[10, 1], sqmea.positions[10, 2], 'y*')
plt.plot(sqmea.positions[-1, 1], sqmea.positions[-1, 2], 'c*')
_ = plt.axis('equal')
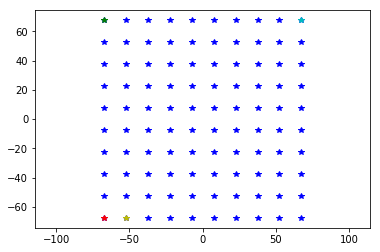
Rectangular MEAs can be handled as matrices, where the first inex is the ROW and the second index is the COLUMN:
print(sqmea[0][0].position) # electrode 0
print(sqmea[9][0].position) # electrode 9
print(sqmea[0][1].position) # electrode 10
print(sqmea[-1][-1].position) # electrode 99
[ 0. -67.5 -67.5]
[ 0. -67.5 67.5]
[ 0. -52.5 -67.5]
[ 0. 67.5 67.5]
Rectangular MEA¶
neuroseeker_info = MEA.return_mea_info('Neuroseeker-128')
pprint(neuroseeker_info)
{'dim': [32, 4],
'electrode_name': 'Neuroseeker-128',
'pitch': 22.5,
'shape': 'square',
'size': 10.0,
'sortlist': None,
'type': 'mea'}
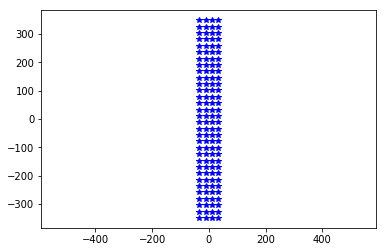
This MEA is rectangular, with 32 rows, 4 columns, and a regular pitch of 22.5um
neuroseeker = MEA.return_mea('Neuroseeker-128')
print(type(neuroseeker))
print(neuroseeker.number_electrodes)
print(neuroseeker.dim)
'plane' field with 2D dimensions assumed to be 'yz
Model is set to semi
<class 'MEAutility.core.RectMEA'>
128
[32, 4]
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'b*')
_ = plt.axis('equal')
print(neuroseeker[0][0].position) # electrode 0
print(neuroseeker[31][0].position) # electrode 31
print(neuroseeker[1][0].position) # electrode 32
print(neuroseeker[-1][-1].position) # electrode 127
[ 0. -33.75 -348.75]
[ 0. -33.75 348.75]
[ 0. -33.75 -326.25]
[ 0. 33.75 348.75]
General MEA¶
When dim and pitch is are single int (or float for
pitch) or a list of 2 values, a rectangular MEA is created. Some MEA
configuration can be different.
neuronexus_info = MEA.return_mea_info('Neuronexus-32')
pprint(neuronexus_info)
{'dim': [10, 12, 10],
'electrode_name': 'Neuronexus-32',
'pitch': [25.0, 18.0],
'shape': 'circle',
'size': 7.5,
'sortlist': None,
'stagger': -12.5,
'type': 'mea'}
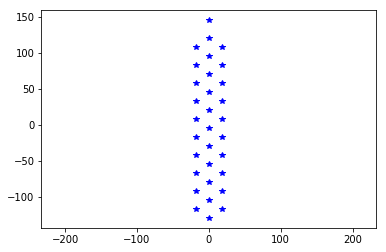
For this MEA there are 3 different options: - dim has 3 elements -
pitch hass 2 elements - stagger is present
When len(dim) > 2, then each element represents the number of rows
of each column. In this case, there are 3 columns: the first and third
have 10 electrodes, the second one has 12.
The first value of pitch is the inter-row distance (top to bottom).
The second value is the inter-column distance (left to right).
The stagger key allows the shift colimns. If only one value is given
(int or float) every other column starting from he second one is
staggered. Otherwise stagger can be a list with the same number of
elements of dim.
Given this information, we can wxpect how the neuronexus MEA looks like:
neuronexus = MEA.return_mea('Neuronexus-32')
plt.plot(neuronexus.positions[:, 1], neuronexus.positions[:, 2], 'b*')
_ = plt.axis('equal')
'plane' field with 2D dimensions assumed to be 'yz
Model is set to semi
Adding and removing MEA models¶
It is possible to load user-defined yaml files in the MEAutility package, so that they are available from the entire file system.
Let’s first create a user.yaml file on-the-fly.
import yaml, os
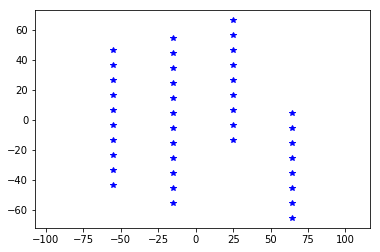
user_info = {'dim': [10, 12, 9, 8],
'electrode_name': 'user',
'description': "a brief description of the probe",
'pitch': [10.0, 40.0],
'shape': 'circle',
'size': 7.5,
'sortlist': None,
'stagger': [0, -12, 30, -22],
'type': 'mea'}
with open('user.yaml', 'w') as f:
yaml.dump(user_info, f)
yaml_files = [f for f in os.listdir('.') if f.endswith('.yaml')]
print(yaml_files)
['user.yaml']
Now we can add the newly created yaml file to the MEA package:
MEA.add_mea('user.yaml')
Available MEA:
['SqMEA-6-25um', 'SqMEA-10-15um', 'tetrode', 'Neuroseeker-128', 'SqMEA-5-30um', 'SqMEA-15-10um', 'Neuronexus-32-Kampff', 'Neuronexus-32-cut-30', 'Neuropixels-128', 'Neuroseeker-128-Kampff', 'Neuropixels-24', 'SqMEA-7-20um', 'Neuronexus-32', 'user', 'Neuropixels-384']
and create a user MEA object:
usermea = MEA.return_mea('user')
plt.plot(usermea.positions[:, 1], usermea.positions[:, 2], 'b*')
_ = plt.axis('equal')
'plane' field with 2D dimensions assumed to be 'yz
Model is set to semi
If we don’t need the user MEA anymore, we can remove it from the MEA
package:
MEA.remove_mea('user')
Removed: /home/alessiob/anaconda3/envs/mearec/lib/python3.6/site-packages/MEAutility/electrodes/user.yaml
Available MEA:
['SqMEA-6-25um', 'SqMEA-10-15um', 'tetrode', 'Neuroseeker-128', 'SqMEA-5-30um', 'SqMEA-15-10um', 'Neuronexus-32-Kampff', 'Neuronexus-32-cut-30', 'Neuropixels-128', 'Neuroseeker-128-Kampff', 'Neuropixels-24', 'SqMEA-7-20um', 'Neuronexus-32', 'Neuropixels-384']