MEA handling¶
This notebook shows how to handle MEA and electrodes in he 3D space.
import MEAutility as MEA
import matplotlib.pylab as plt
First, let’s instantiate a MEA object among the available MEAs:
MEA.return_mea()
Available MEA:
['SqMEA-6-25um', 'SqMEA-10-15um', 'circle_500', 'tetrode', 'Neuroseeker-128', 'SqMEA-5-30um', 'SqMEA-15-10um', 'Neuronexus-32-Kampff', 'Neuronexus-32-cut-30', 'Neuropixels-128', 'Neuroseeker-128-Kampff', 'Neuropixels-24', 'SqMEA-7-20um', 'Neuronexus-32', 'Neuropixels-384']
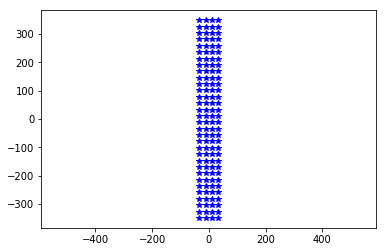
neuroseeker = MEA.return_mea('Neuroseeker-128')
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'b*')
_ = plt.axis('equal')
By default the MEA is instantiated with it’s center of mass at (0,0,0)
and electrodes lying in the plane specified in the yaml file (by default
plane is yz)
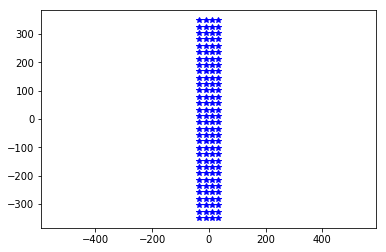
neuroseeker.plane
'yz'
Moving the probe around¶
The probe can be easily moved with a the move and center
methods:
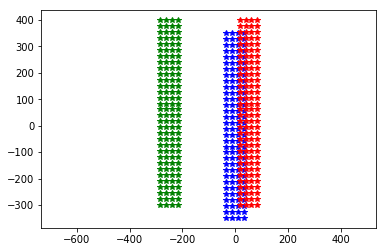
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'b*')
neuroseeker.move([0, 50, 50])
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'r*')
neuroseeker.move([0, -300, 0])
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'g*')
_ = plt.axis('equal')
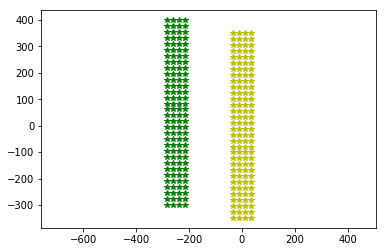
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'g*')
neuroseeker.center()
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'y*')
_ = plt.axis('equal')
Rotating the probe¶
With the rotate method, MEA probes can be rotated along any axis by
any angle (in degrees). The current plane and orientation of the probe
is stored by the variables main_axes and normal
# main_axes indicate the MEA plane
print(neuroseeker.main_axes[0], neuroseeker.main_axes[1])
# normal indicates the axis perpendicular to the electrodes
print(neuroseeker.normal)
# normal axis is also stored by each electrode and could be changed separately
print(type(neuroseeker.electrodes[0]), neuroseeker.electrodes[0].normal)
[0 1 0] [0 0 1]
[-1. 0. 0.]
<class 'MEAutility.core.Electrode'> [-1. 0. 0.]
Now le’s make some rotations!!
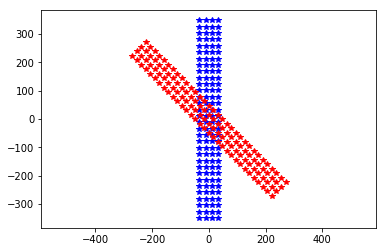
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'b*')
neuroseeker.rotate([1, 0, 0], 45)
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'r*')
_ = plt.axis('equal')
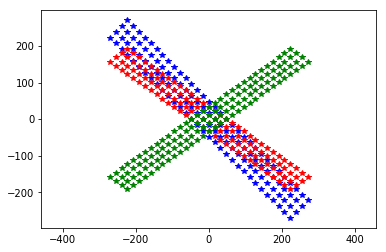
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'b*')
neuroseeker.rotate([0, 1, 0], 45)
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'r*')
neuroseeker.rotate([0, 1, 0], 90)
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'g*')
_ = plt.axis('equal')
# back to normal
neuroseeker.rotate([0, 1, 0], -90)
neuroseeker.rotate([0, 1, 0], -45)
neuroseeker.rotate([1, 0, 0], -45)
plt.plot(neuroseeker.positions[:, 1], neuroseeker.positions[:, 2], 'b*')
_ = plt.axis('equal')